Introduction
Oral cancer is the sixth most common malignancy worldwide and is particularly prevalent in developing countries, such as in Southeast Asia, where up to 40 percent of all malignancies are located within the oral cavity. More than 90% of cancers in the mouth are Squamous cell carcinomas (SCCs) originating from the oral mucosa. With an average all stage, 5-year survival rate for oral cancer of less than 50%, the annual mortality figures are comparable to those of carcinoma of the cervix and malignant melanoma. In the past, oral cancer predominantly affected men in their sixth or seventh decade. However, more recently the male-to-female ratio has reduced dramatically and there has been a striking increase in the number of cases in those under the age of 45.[1]
Biological Microflora
Despite the widening interest in the possible association between bacteria and different stages of cancer development, our knowledge in its relation to oral cancers remains inadequate. Different bacteria have been proposed to induce carcinogenesis either through induction of chronic inflammation or by interference, either directly or indirectly, with eukaryotic cell cycle and signaling pathways, or by metabolism of potentially carcinogenic substances like acetaldehyde causing mutagenesis. Studies have shown diversity of isolated bacterial taxa between the oral cancer tissue specimens and the control, with Exiguobacterium oxidotolerans, Prevotella melani nogenica, Staphylococcus aureus and Veillonella parvula being specific for tumorogenic tissues. Most isolates are saccharolytic and acid tolerant. Streptococcus anginosus, commonly linked with esophageal and pharyngeal cancers, is not of significance in oral cancers. Similarly, significant salivary specificity is noted for three bacteria, namely, Capnocytophaga gingivalis, P. melaninogenica, and Streptococcus mitis in oral cancer patients, making these species salivary markers for the early detection of oral cancers and thus improving the survival rate significantly. Also, such high degree of bacterial specificity in oral cancers has provoked the designing of new treatment options for cancer prevention by way of vaccine delivery. However, for the success of these steps, a deeper exploration into this subject with a greater understanding is warranted.[2]
Interest in the possible relationships between bacteria and the different stages of cancer development has been increasing since the classification by the World Health Organization of Helicobacter pylori as a definite (Class 1) carcinogen. Various other bacterial infections have also been found to correlate with an increased risk of developing cancer, for instance, an increased risk of gallbladder carcinoma is associated with Salmonella typhi infection and there is a greater risk of developing colon cancer in Streptococcus bovis-infected patients.[3]
History: Associated Etiological Bacteria
Several discoveries in microbiological literature since 19th century have led its way to suggest that bacteria were implicated in all studies, and hence, the theory of bacterial infection leading to oral cancer was born. Various epidemiological and laboratory based studies have shown number of bacterial species to be associated with different cancers. Few such propositions that gained widespread interest were following revelations.[4]
1. Salmonella typhi and gallbladder cancer
2. Chlamydophila pneumoniae and lung cancer
3. Streptococcus bovis and colorectal cancer
4. E. coli, crohn's disease and colon cancer
5. S. typhi and susceptible populations[5]
Pathogenesis:
The pathogenesis of carcinogenesis due to bacteria can be attributed to two mechanisms viz:
a) Chronic inflammatory mechanisms
b) Role of Bacterial toxins
Bacteria and Oral Cancer
The association of bacteria with oral tumors is of increasing interest. In a study of intraoral carcinomas, Nagy et al. (1998) demonstrated a difference in the microflora associated with the surface of tumors in comparison to control sites. More recently, it has also been reported that patients with oral squamous cell carcinoma (OSCC) tend to possess significantly raised concentrations of certain bacteria in their saliva. In order to demonstrate a role for bacteria in the development of oral cancer, the first step must be to identify such organisms within tumor specimens. Furthermore, sufficient attention must be given to the elimination from any tissues tested of the microbes that occur naturally on the surfaces of the tumors. In addition, salivary contamination of the sample must be prevented during subsequent handling. The presence of Streptococcus anginosus DNA in oropharyngeal tumors has been reported following studies using specific PCR primers. However, this molecular approach was limited to a single group of bacteria, and no inferences can be made regarding the viability and therefore potential activity of the species detected.[5] To date there have been only a few investigations into the possible associations between bacterial species and oral carcinoma. In a study of intraoral carcinomas, Nagy et al. (1998) demonstrated increased numbers of certain members of the oral microbiota on the surface of tumors in comparison with control sites. More recently, it has been reported that patients with OSCC tend to possess significantly raised concentrations of certain bacteria in their saliva. This apparent alteration of the oral microbiota in cases of OSCC is of particular interest because of its potential application as a diagnostic tool.[7]
Bacterial tropism in oral cavity
Research has repeatedly shown that oral bacteria demonstrate specific tropisms toward different biological surfaces in the oral cavity such as the teeth, mucosa, and other bacteria. The reason for these shifts in bacterial colonization of cancer lesions is unclear. Mechanistic studies of bacterial attachment provide some insights, however. The non-shedding surfaces of the teeth offer a far different habitat than the continually shedding surfaces of the oral mucosa.[8]
Due to the repeated shedding of epithelial cells, there is less time for a complex biofilm to develop on soft tissue surfaces; thus, a premium is placed on potent mechanisms of adhesion. The differences in bacterial tropisms for specific oral sites suggest that different intra-oral surfaces and bacterial species have different receptors and adhesion molecules that dictate the colonization of different oral surfaces.[9]
It is now recognized that bacteria bind to and colonize mucosal surfaces in a highly selective manner via a "lock and key" mechanism.[10] Adhesins on bacteria bind specifically to complementary receptors on the mucosal surfaces of the host.[11] These adhesins differ from species to species leading to specificity in attachment to different surfaces.[12] Studies have shown that even within genera, colonization patterns of individual species may differ markedly Streptococcus salivarius, for example, preferentially colonized the oral soft tissues and saliva compared to the teeth, while the reverse was true of Streptococcus sanguis.[13]
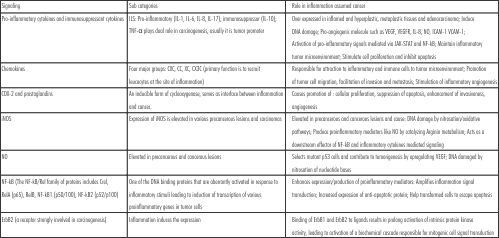 | a. Chronic inflammatory mechanisms involved in carcinogenesis:
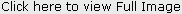 |
 | b. Role of Bacterial toxins in Carcinogenesis
 |
Diagnostic Modalities
Oral squamous cell carcinoma (OSCC) is one of the most common epithelial malignancies with significance morbidity and mortality. In spite of diagnostic and therapeutic advances over the decades, the disease still remains a challenge for medical professionals with the five year survival rate being 30%-50%.[14] An understanding of the molecular mechanisms involved in OSCC is helpful in providing a more complete picture of the ways in which tumor arise and advance and a rationale for novel strategies of cancer detection.[15] The oral cavity is particularly conducive to such strategies, given the ease with which saliva and exfoliated cells can be collected.[16] Tumor cells inhabit or produce biochemical substances referred to as tumor markers. These can be normal endogenous products that are produced at a greater rate in cancer cells or the products of newly switched on genes that remain quiescent in the normal cells.[17] Tumor marker may be present as intracellular substances in tissues or as released substances in circulating body fluids such as serum, urine, cerebrospinal fluid (CSF) and saliva. Until recently, analysis for tumor markers were carried out in fluids other than saliva such as CSF, blood and urine.[18] With recent diagnostic technological advances however, the role of saliva as a tool for
diagnosis has advanced exponentially. The source of information is largely derived from the variety of DNA's, RNA's and proteins present in the saliva. Salivary DNA represents the genetic information of the hosting human body, the oral microbiota and the infecting DNA-viruses. Salivary RNA provides information on the transcription rates of the host genes and those of oral microbiota. Salivary proteins represent genetic information and help to understand the translational regulation of the host body and the oral microbiota.[17] In addition, saliva is also useful in detection of other markers such as cell cycle markers (p16, p53 etc), growth factors (epidermal growth factor, transforming growth factor etc), cell surface markers, oxidant and antioxidants among others.[19]
Technologies for saliva based diagnosis
1. Proteomics
The proteome represents the complete set of proteins encoded by genome and proteomics is the study of the proteome that investigates the cellular levels of all the isoforms and post translational modifications of proteins that are encoded by the genome of the cell under a given set of circumstances.[20]
Protein biomarkers in saliva are being analyzed both individually and as a panel of markers to aid in early detection of oral cancer and in implementing appropriate therapeutic regime[21]
2. Transcriptomics
Salivary transcriptome diagnostics constitutes a novel clinical approach where a large panel of human RNAs can be readily detected in saliva. The large panel of human mRNA is determined by the use of microarray technology and after profiling, validation of transcriptome biomarkers is done by quantitative real time PCR. Sometimes multiplex reverse transcriptase PCR are used to overcome problems with quantitative real time PCR. Microarray technologies have the advantage of simultaneously detecting and quantifying the expression of large number of genes health and disease. The technology involves the use of robotic automated miniaturized microscopic spots of aliquots of cDNAs or oligonucleotides from specific genes in a standardized high density gridded arrangement on glass.[22]
3. Polymerase chain reaction (PCR)
PCR is a simple in-vitro method for amplification of specific short segments of DNA or cDNA reverse transcribed from RNA. The technique greatly simplifies genetic analysis and permits the study of all types of clinical samples. Oligonucleotide primers binding to the flanking regions of target sequence are used to initiate specific copying of DNA strands by DNA polymerase.[23] The requirement for the reactions are template DNA to be studied, short single strand DNA primers, complementary to opposite strands of the flanking regions of the fragment of interest, the four nucleotide triphosphatases, a thermostable DNA polymerase and an appropriate buffer solution.[24]
4. Genomics
Genomic analysis is one of the recent advances in the diagnosis of oral cancer and considerable research is being performed in this field. With the availability of high throughout technologies to harness genetic information from various sources like blood, saliva, etc., their usage has advanced exponentially. Stable cell free circulating DNA in plasma was first observed almost 60yrs ago. Plasma DNA were shown to exhibit tumor specific characteristics such as somatic mutations in tumor suppressor genes or oncogenes, microsatellite alterations, abnormal protein methylation, mitochondrial DNA mutations and presence of tumor related viral RNA. These DNA related changes were also found in saliva, the identification of which helps a great deal in diagnosis of oral cancer.[25]
Tumor specific genomic markers consisting of DNA and RNA markers can be identified in saliva for detection of oral cancer considering that the initiation and progression of malignant tumors is driven by the accumulation of specific genetic alterations.[26]
Future Perspectives and Conclusion
The challenge of understanding the true associations between bacterial infections and human cancers is indeed great, but it also promises great rewards. Unlike viral infections, bacterial infections are typically curable, and the prospect of antibiotic treatments to prevent, alleviate, or cure cancers is obviously alluring. Many of the bacterial infections that promote oncogenesis, i.e., H. pylori, Chlamydia, and Mycoplasma infections, are often asymptomatic. When the pathways toward malignancy are initiated and when they become irreversible, though, are not fully understood. Vaccination against etiologic pathogens to prevent infection and thus eliminate the risk of cancer is yet another hopeful prospect for researchers.[27]
One of the most intimate relationships of man is that which he has with his own microbial flora.[28] While, most exposures in life are transient, the contact we have with these microorganisms is constant and unremitting. This symbiotic relationship is taken for granted or, more commonly still, thought to be beneficial.[29] Even the term normal flora suggests benignity yet, it is naïve to assume that our continuous interaction with microbial flora is immaterial to our long-term health. As new infectious causes of malignancy continue to be uncovered, it is increasingly apparent that dissection of the complex interplay between man and microbial flora is essential to understanding the pathogenesis of many malignancies.
Recent research has uncovered a great deal of information regarding the bacterial mechanisms used to cause, colonize or cure cancer, however, many questions remain unanswered. The continued exploration of the relationship between bacteria and oral squamous cell carcinoma will bring research ever closer to the prevention, early diagnosis and truly effective treatment of this scourge of mankind.[1]
References
1. Hooper SJ, Crean SJ, Lewis MAO, Spratt DA, Wade WG, Wilson MJ. Viable Bacteria Present within oral squamous cell carcinoma tissue. J Clin Microbiol (2006), 44(5): 1719-1725.
2. Pushalkar S, Mane SP, Ji X, Li Y, Evans C, Crasta OR et al. Microbial Diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol (2011), 61(3): 1-9.
3. Hooper SJ, Crean SJ, Fardy MJ, Lewis MAO, Spratt DA, Wade WG et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol (2007), 56: 1651-1659.
4. Chocolatewala N, Chaturvedi P, Desale R. The role of bacteria in oral cancer. J Med Microbiol (2007), 31(4): 126-131.
5. DL Mager. Bacteria and cancer: cause, coincidence or cure? A review. Journal of Translational Medicine 2006, 4:14.
6. Nath G, Gulati AK, Shukla VK. Role of bacteria in carcinogenesis, with special reference to carcinoma of the gallbladder. World J Gastroenterol (2010), 16(43): 5395-5404.
7. Kenna GA. Bacteria, oral cancer connection reported. JADA (2005), 136: 1222.
8. Bahar G, Feinmesser R, Popovtzer A, Nagler RM. Salivary analysis in oral cancer patients: DNA and protein oxidation, reactive nitrogen species and anti oxidants profile. Cancer (2006), 109(1): 54-59.
9. Miyazaki Y, Kikuchi K, Gonzalez-Alva P, Inoue H, Noguchi Y, Tsuchiya H, et al. Association of butyric acid produced by periodontopathic bacteria with progression of oral cancer. J Cancer Sci Ther (2010), 2(2): 26-32.
10. Krasse B. The proportional distribution of Streptococcus salivarius and other streptococci in various parts of the mouth. Odontol Revy (1954) 5(3): 203-211.
11. van Houte J, Gibbons RJ, Banghart SB. Adherence as a determinant of the presence of Streptococcus salivarius and Streptococcus sanguis on the human tooth surface. Arch Oral Biol (1970), 15:1025-1034.
12. Liljemark WF, Gibbons RJ. Proportional distribution and relative adherence of Streptococcus miteor (mitis) on various surfaces in the human oral cavity. Infect Immun 1972, 6(5): 852-859.
13. Van Houte J, Gibbons RJ, Pulkkinen AJ. Adherence as an ecological determinant for streptococci in the human mouth. Arch Oral Biol (1971), 16(10): 1131-1141.
14. Chiappin S, Antonelli G, Gatti R, De Palo EF. Saliva Specimen: A new laboratory tool for diagnostic and basic investigation. Clin Chim Acta (2007); 383(1-2): 30-40.
15. Franzmann EJ, Reategui EP, Pedroso F, Pernas FG, Karakullukcu BM, Carraway KL. Salivary soluble CD44: a potential marker for head and neck cancer. Cancer Epidemiol Biomarkers Prev (2005), 14(3): 735-739.
16. Holm-Hansen C, Tong G, Davis C, Abrams WR, Malamud D. Comparison of oral fluid collectors for use in point-of-care diagnostic device. Clin Diagn Lab Immunol (2004), 11(5): 909-912.
17. Hu S, Denny P, Denny P, Xie Y, Loo JA, Wolinsky LE. Differentially expressed protein markers in human submandibular and sublingual secretions. Int J Oncol (2004), 25(5): 1123-1140.
18. Kaufmann E, Lamster B. The diagnostic applications of saliva-A review. CROBM (2002), 13: 197-212.
19. Liao PH, Chang YG, Huang MF, Tai KW, Chou MY. Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinoma. Oral Oncol (2000); 36(3): 272-276.
20. Malati, T. Tumor markers: An overview. Ind J Clin Biochem (2007), 22(2): 17-31.
21. Streckfus CF, Bigler LR. Saliva as a diagnostic fluid. Oral Dis (2002), 8: 69-76.
22. Mandel ID. The diagnostics uses of saliva. J Oral Pathol Med (1990), 19: 119-125.
23. Tenevuo JO. Human saliva. Clinical chemistry and microbiology. Studies in Stomatology & Craniofacial Biology, CRC Press Inc (1989), Boca Raton.
24. Muelker RF, Young DI. Emery's elements of medical genetics. Churchill Livingstone; Exeter, UK, 12th Edition.
25. Rai, B, Kharb S, Jain R, Anand SC. Salivary lipid peroxidation product malonaldehyde in pre-cancer and cancer. Adv Med Dent Sci (2008), 2(1): 7-8.
26. Sun Y. Free radicals, antioxidant enzymes and carcinogenesis. Free Radic Biol Med (1990), 8: 583-599.
27. Chang AH, Parsonnet J. Role of bacteria in oncogenesis. Clin Microbiol Rev (2010), 23(4): 837-857.
28. Dayama A, Srrivastava V, Shukla M, Singh R, Pandey M. Helicobacter pylori and oral cancer: Possible association in a preliminary case control study. Asian Pacific J Cancer Prev (2011), 12: 1333-1336.
29. Parsonnet J. Bacterial infection as a cause of cancer. Environ Health Perspect (1995), 103(8): 263-268. |